Baojin Ding

2001 B.S. (Medicine, MD equivalent) Medical College of Qingdao University, China
2004 M.S. (Clinical Laboratory) Wenzhou Medical University, China
2010 Ph.D. (Biochemistry and Molecular Biology) Louisiana State University, Baton Rouge LA
Our research focuses on cellular and molecular neuroscience and neurological diseases. Cell culture and genetically modified mice are used as model systems. Techniques in Molecular Biology, Cell Biology and Biochemistry are employed. Our research projects can be grouped into three closely correlated directions.
1. The timing mechanism of gene expression in maturing neurons
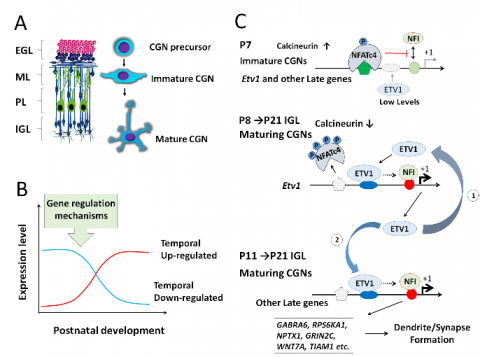
Central nervous system development results from the interactions between intrinsic genes and extrinsic environment. The process of neuronal development consists of successive developmental stages including proliferation, differentiation, migration, ax
on extension, dendritogenesis, and formation of functional synapses. In this developmental sequence, the interplay between genes and environment ensures that each step must occur in the proper timing and sequence. Their successful regulation requires that numerous groups of genes be turned on and off in a timely manner. However, the timing mechanisms of gene expression in maturing neurons are not fully understood.
By using mouse cerebellar granule neurons (CGNs) as a research model, we have identified a novel nuclear factor one (NFI)-regulated temporal switch program linked to dendrite formation. In this program, neuronal mature genes (late expressed synaptoge nesis-related genes) are up-regulated and immature genes (early expressed amplification-related genes) are down-regulated during a period of postnatal development window. One distinguished feature of this program is the NFI temporal occupancy of target genes. We also found that NFI switch program were regulated by resting membrane potential, CaN/NFAT signaling pathway, ETV1 and BDNF etc. Most interestingly, some neurodevelopmental disorders are associated with particular NFI-regulated 'switch' genes, such as Autism Spectrum Disorders (ASD). Research efforts are focusing on identification and characterization of novel regulators and signaling pathways of NFI-regulated program, and the involvement of disrupted NFI-regulated developmental program in neurodevelopmental disorders.
2. Nucleocytoplasmic transport regulation
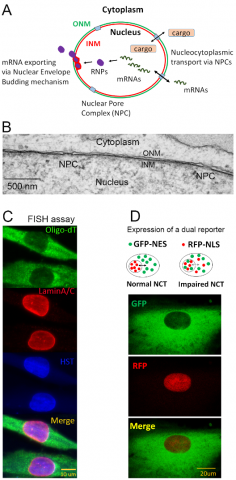 In eukaryotic cells, transcription and translation processes are physically separated by nuclear envelope (NE). Thus, the proper functions of a cell involves the mRNA nuclear exporting for protein synthesis in the cytoplasm and protein nuclear transport cross the NE (Figure 2A). Defective nucleocytoplasmic transport (NCT) has merged as a common molecular underpinning of many neurological diseases, including Alzheimer disease (AD), amyotrophic lateral sclerosis (ALS) and Huntington’s disease (HD). Systematically analyze the regulation of nucleocytoplasmic transport will provide novel insights into the nuclear transport machinery and advance our understanding of how defective NCT contributes to human diseases. We use electron microscope (EM) to examine the ultrastructure of cells, including nuclear envelope and nuclear pore complex (NPC). We have established two systems to analyze nuclear transport function in cultured mammalian cells. One approach measures nuclear mRNA exporting using fluorescent in situ hybridization (FISH) with oligo-dT probes combined with immuno-staining of NE markers. Another approach analyzes protein nuclear transport using lentiviral delivery of a dual reporter (GFP-NES and RFP-NLS). Research efforts are focusing on molecular mechanisms of NCT machinery and identification of dysregulated factors that disrupt NCT in aging and diseased cells.
In eukaryotic cells, transcription and translation processes are physically separated by nuclear envelope (NE). Thus, the proper functions of a cell involves the mRNA nuclear exporting for protein synthesis in the cytoplasm and protein nuclear transport cross the NE (Figure 2A). Defective nucleocytoplasmic transport (NCT) has merged as a common molecular underpinning of many neurological diseases, including Alzheimer disease (AD), amyotrophic lateral sclerosis (ALS) and Huntington’s disease (HD). Systematically analyze the regulation of nucleocytoplasmic transport will provide novel insights into the nuclear transport machinery and advance our understanding of how defective NCT contributes to human diseases. We use electron microscope (EM) to examine the ultrastructure of cells, including nuclear envelope and nuclear pore complex (NPC). We have established two systems to analyze nuclear transport function in cultured mammalian cells. One approach measures nuclear mRNA exporting using fluorescent in situ hybridization (FISH) with oligo-dT probes combined with immuno-staining of NE markers. Another approach analyzes protein nuclear transport using lentiviral delivery of a dual reporter (GFP-NES and RFP-NLS). Research efforts are focusing on molecular mechanisms of NCT machinery and identification of dysregulated factors that disrupt NCT in aging and diseased cells.
3. Modeling human neurological diseases
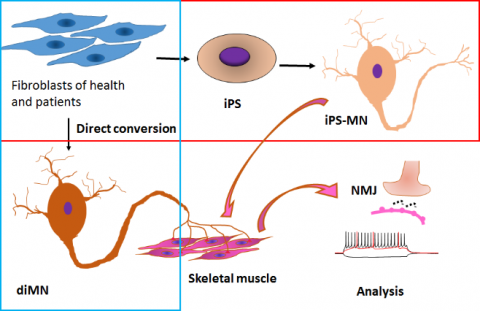 The limited access to patient neurons greatly impedes the progress of research in neurological diseases. Reprogramming of human neurons from adult fibroblasts provides an unprecedented approach to deciphering the molecular pathogenesis underlying disease condition. Using lentiviral delivery of transcription factors, we can generate human neurons from adult fibroblast via two strategies. One strategy is direct conversion of fibroblasts into neurons. The other strategy is induced pluripotent stem cells (iPSCs)-based reprogramming and differentiation. Research efforts are focusing on the molecular pathogenesis of neurological disorders, such as Alzheimer disease (AD), Dystonia and amyotrophic lateral sclerosis (ALS).
The limited access to patient neurons greatly impedes the progress of research in neurological diseases. Reprogramming of human neurons from adult fibroblasts provides an unprecedented approach to deciphering the molecular pathogenesis underlying disease condition. Using lentiviral delivery of transcription factors, we can generate human neurons from adult fibroblast via two strategies. One strategy is direct conversion of fibroblasts into neurons. The other strategy is induced pluripotent stem cells (iPSCs)-based reprogramming and differentiation. Research efforts are focusing on the molecular pathogenesis of neurological disorders, such as Alzheimer disease (AD), Dystonia and amyotrophic lateral sclerosis (ALS).
Selected Publications:
- Ding B, Dobner PR, Mullikin-Kilpatrick D, Wang W, Zhu H, Chow CW, Gronostajski RM and Kilpatrick DL. (2018). BDNF Activates an NFI-Dependent Neurodevelopmental Timing Program By Sequestering NFATc4. Mol Biol Cell. 29(8):975-987
- Ding B., Mirza A,M., Alshley J. Budnik V. and Munson M. (2017) Nuclear Export Through Nuclear Envelope Remodeling in Saccharomyces cerevisiae. (bioRxiv 224055; doi: https://doi.org/10.1101/224055)
- Leto K, Arancillo M, Becker EB, Buffo A, Chiang C, Ding B, Dobyns WB, Dusart I, Haldipur P, Hatten ME, Hoshino M, Joyner AL, Kano M, Kilpatrick DL, Koibuchi N, Marino S, Martinez S, Millen KJ, Millner TO, Miyata T, Parmigiani E, Schilling K, Sekerková G, Sillitoe RV, Sotelo C, Uesaka N, Wefers A, Wingate RJ, Hawkes R. (2016) Consensus Paper: Cerebellar Development. Cerebellum. Dec;15(6): 789-828.
- Li Y, Hassinger L, Thomson T, Ding B, Ashley J, Hassinger W and Budnik V. (2016). Lamin Mutations Accelerate Aging via Defective Export of Mitochondrial mRNAs through Nuclear Envelope Budding. Curr Biol. 26(15):2052-2059
- Ding B, Cave HW, Dobner PR, Kilpatrick DM, Bartsokis M, Zhu H, Chow CW, Gronostajski RM and Kilpatrick DL. (2016) Reciprocal Auto-Regulation by NFI Occupancy and ETV1 Promotes the Developmental Expression of Dendrite-Synapse Genes in Cerebellar Granule Neurons. Mol Biol Cell. 27(9):1488-99
- Ding B. (2015) How does a 1.5-Fold Increase in Gene Dosage in Chromosome 21 Cause the Pleiotropic Phenotypes in Down Syndrome? J Down Syndr Chr Abnorm 1: 1: e101. doi:10.4172/jdsca.1000e101 (Editorial)
- Packard M, Jokhi V, Ding B and Budnik V. (2015) Nucleus to Synapse Nesprin Railroad Tracks Direct Synapse Maturation through RNA localization. Neuron. 86(4):1015-28).
- Ding B. (2015) Gene Expression in Maturing Neurons: Regulatory Mechanisms and Related Neurodevelopmental Disorders. ACTA PHYSIOLOGICA SINICA. 67(2):113-33. (Invited Review)
- Ding B, Wang W, Selvakumar T, Xi HS, Zhu H, Chow CW, Horton JD, Gronostajski RM and Kilpatrick DL. (2013) Temporal Regulation of Nuclear Factor One Occupancy by Calcineurin/NFAT Governs a Voltage-Sensitive Developmental Switch in Late Maturing Neurons. J Neurosci. 33(7):2860-2872.
- Ding B and Kilpatrick DL. (2013) Lentiviral Vector Production, Titration, and Transduction of Primary Neurons. Methods Mol Biol. 1018:119-31. Chapter 12.
- Ding B and Kilpatrick DL. Kilpatrick (2013). Chromatin Immunoprecipitation Assay of Brain Tissue Using Percoll Gradient-Purified Nuclei. Methods Mol Biol. 1018:199-209. Chapter 19.
- Ding B, Lejeune D and Li S. (2010) The C-terminal Repeat Domain of Spt5 Plays an Important Role in Suppression of Rad26-independent Transcription Coupled Repair. J Biol Chem. 285 (8): 5317-5326.
- Chen X, Ding B, Lejeune D, Ruggiero C and Li S. (2009) Sumoylation of Rpb1 in Response to UV Radiation or Impairment of Transcription Elongation in Yeast. PLoS One. 4 (4) e5267.
- Ding B, Ruggiero C, Chen X and Li S. (2007) Tfb5 is Partially Dispensable for Rad26 Mediated Transcription Coupled Nucleotide Excision Repair in Yeast. DNA Repair (Amst). 6 (11): 1661- 1669.
- Li S, Ding B, LeJeune D, Ruggiero C, Chen X and Smerdon MJ. (2007) The Roles of Rad16 and Rad26 in Repairing Repressed and Actively Transcribed Genes in Yeast. DNA Repair (Amst). 6 (11): 1596-1606.
- Li S, Ding B, Chen R, Ruggiero C and Chen X. (2006) Evidence that Transcription Elongation Function of Rpb9 is Involved in Transcription Coupled DNA Repair in Saccharomyces cerevisiae. Mol Cell Biol. 26 (24): 9430-9441.
- Ding B, Smith ES, and Ding H. (2005) Mobilization of Iron Center in IscA for Iron-sulfur Cluster Assembly in IscU. Biochem. J. 389 (Pt 3):797-802.
More publications: https://www.ncbi.nlm.nih.gov/pubmed/?term=Baojin+Ding
Website: DingLabBioMed.com
Positions
Our lab is currently recruiting highly motivated graduate students and postdoctoral fellows. To apply, please contact Dr. Baojin Ding.
Baojin.Ding@Louisiana.edu, Telephone: (337) 482-1101
